CSIR-CIMAP Bioinformatics Tools and Databases
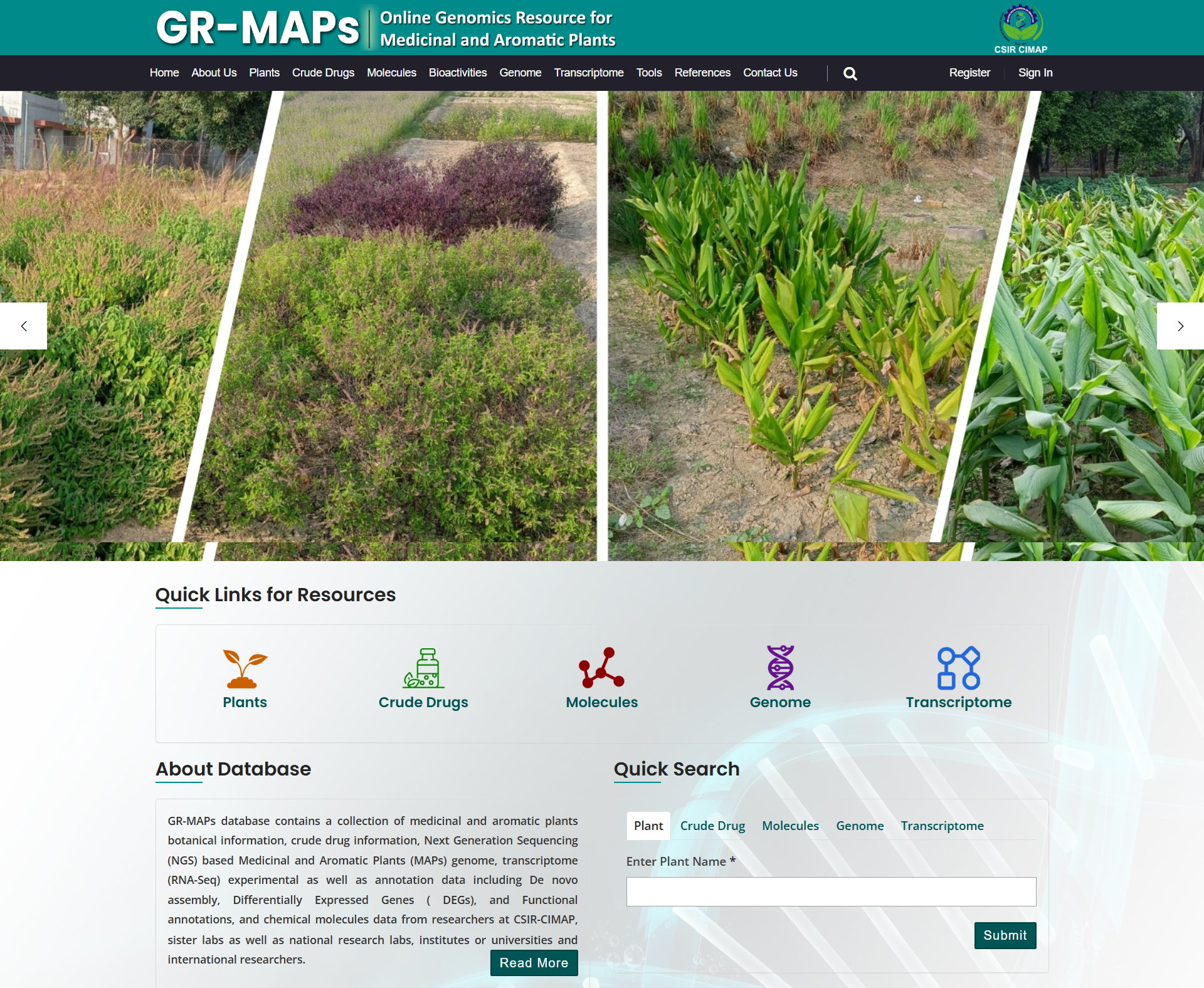
GR-MAPs database contains a collection of medicinal and aromatic plants botanical information, crude drug information..
Read More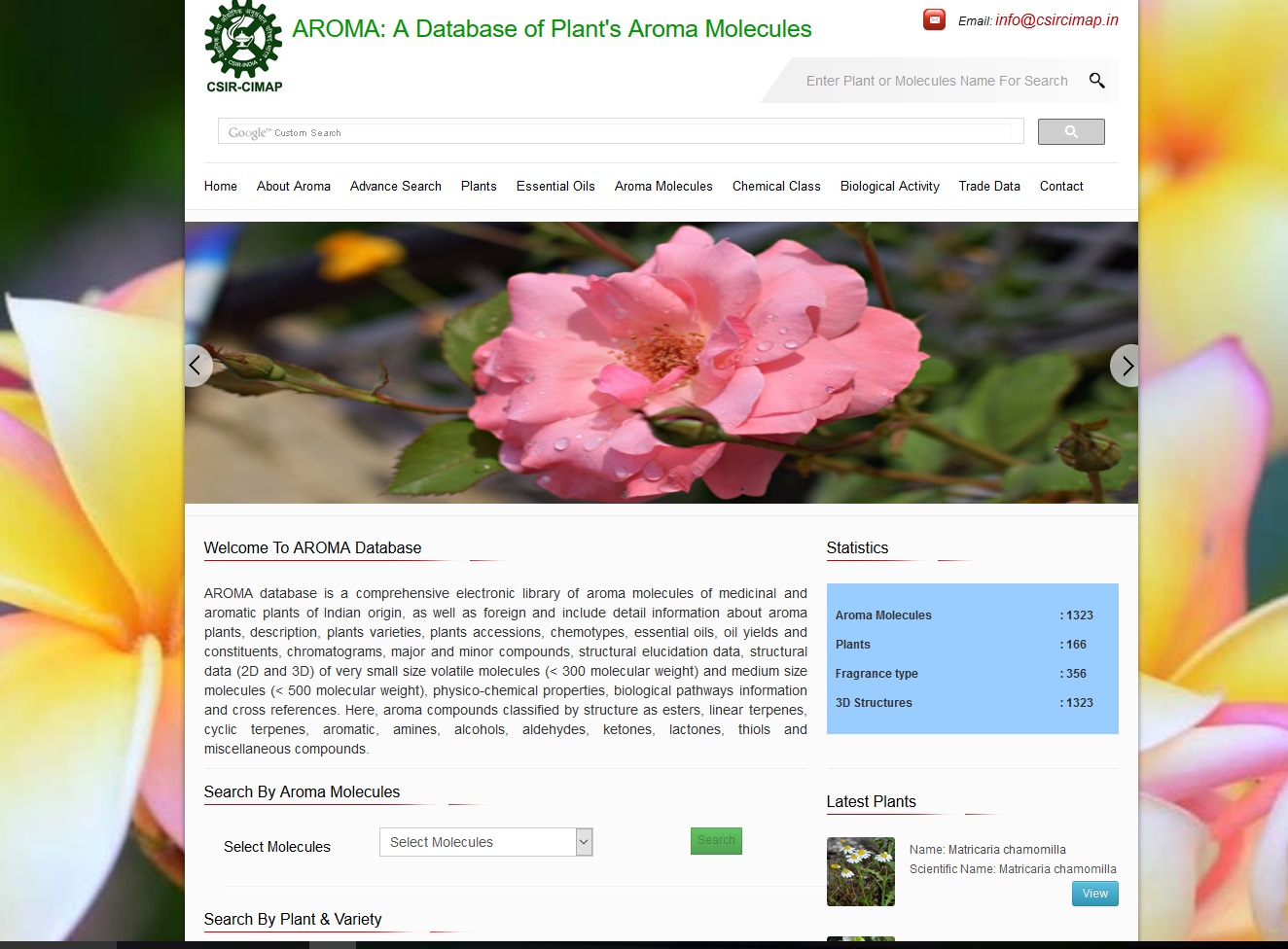
AROMA database is a comprehensive electronic library of aroma molecules of medicinal and aromatic plants of Indian origin.
Read More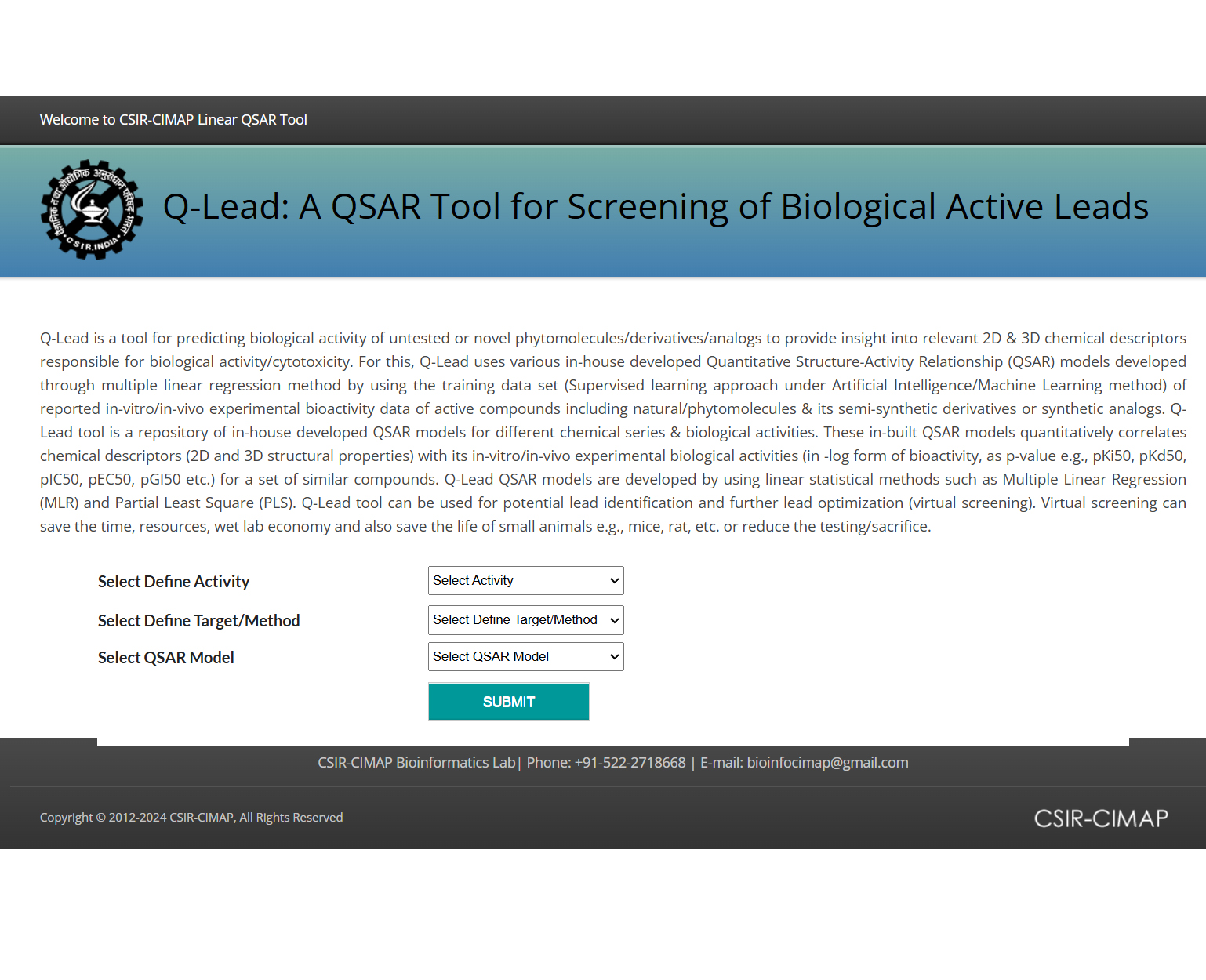
The Q-Lead is a software tool for predicting biological activity of untested natural molecules and it's derivatives/analogues
Read MoreMore information about Databses and Tools
GR-MAPs DatabaseA Database contains a collection of medicinal and aromatic plants botanical information
GR-MAPs database contains a collection of medicinal and aromatic plants botanical information, crude drug information, Next Generation Sequencing data of Medicinal and Aromatic Plants (MAPs) such as genome, transcriptome (RNA-Seq) experimental as well as annotation data including Reference genome/transcriptome (pooled data) based or De-novo assembly data, QC details, Differentially Expressed Genes (DEGs), and Functional annotation details (Homology, Protein family/domain/motif details, Gene Onotology (GO) parameters details, CYPs database match results, KEGG Pathways details, and other Enrichment studies data). This comprehensive MAPs genomic resource database also covered details of crude drugs, phytochemicals & their derivatives with bioactivity details and references.
AROMA DatabaseA Database of Plants based Aroma Molecules
AROMA database is a comprehensive electronic library of aroma molecules of medicinal and aromatic plants of Indian origin,
as well as foreign and include detail information about aroma plants, description, plants varieties, plants accessions, chemotypes,
essential oils, oil yields and constituents, chromatograms, major and minor compounds, structural elucidation data, structural data
(2D and 3D) of very small size volatile molecules (< 300 molecular weight) and medium size molecules (< 500 molecular weight),
physico-chemical properties, biological pathways information and cross references. Here, aroma compounds classified by structure as esters,
linear terpenes, cyclic terpenes, aromatic, amines, alcohols, aldehydes, ketones, lactones, thiols and miscellaneous compounds.
Publication: Kumar Y, Prakash O, Tripathi H, Tandon S, Gupta MM, Rahman LU, Lal RK, Semwal M, Darokar MP, Khan F. AromaDb:
A Database of Medicinal and Aromatic Plant's Aroma Molecules With Phytochemistry and Therapeutic Potentials. Frontiers in plant science. 2018;9.
(PMID: 30150996)
A QSAR Modeling based Screening Tool for Active Leads Prediction.
Summary: The Q-Lead is a software tool for predicting biological activity of untested natural molecules and it's derivatives/analogues, to provide molecular insight into relevant 2D & 3D chemical descriptors responsible for certain biological activity. For this, Q-Lead uses various in-house developed local and global Quantitative Structure-Activity Relationship (QSAR) models based on different natural chemical series. The Q-Lead models quantitatively correlate chemical descriptors (structural properties) with biological activities for a set of similar compounds by using statistical modeling such as Multiple linear Regression (MLR) or PLS. The Q-Lead tool covers 2D QSAR models, which mimicks the bioactivity evaluation through in vitro studies, including both cell line and target based screening assays. The QSAR models included chemical series of following natural compounds e.g., 18ß-Glycyrrhetinic acid (GA), Ursolic acid (UA), Camptothecin (CPT), Chalcone, artemisinin, vaniloids (capsaicin and capsazepine), and Xanthone derivatives for anticancer, antibacterial, antimalarial and immunomodulatory/antiinflammamtrory activities, both cell line and target enzyme specific. The Q-Lead tool could be more useful in early identification of potential drug-like leads.